Epigenetic Clocks are No Longer Just for Humans
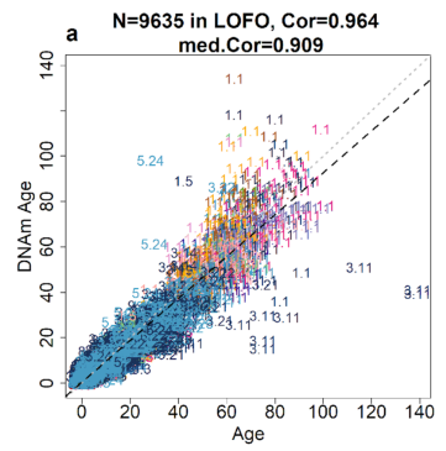
Over the last several years, with generous support from the Paul G. Allen Frontiers Group, Dr. Steve Horvath at UCLA has profiled DNA methylation for over 125 different mammalian species, including dogs, cats, elephants, horses, primates, bats, mice, rats, naked mole rats, cattle, sheep, pigs, bowhead, beluga, and killer whales, bottlenose dolphins, and many more. Remarkably, as part of this work, it was discovered that a universal mammalian clock could be generated, which accurately predicts chronological age across all mammalian species. Many different tissues were analyzed including blood, liver, kidney, muscle, skin, and the brain, including many different regions of it (cerebellum, cortex, hippocampus, hypothalamus, striatum, and subventricular zone). Universal clocks generated based on this data use a single mathematical formula and are highly accurate age estimators (r>0.96).
The work also enabled the development of the Horvath Mammalian Array, which addresses the fact that it is difficult to develop custom methylation arrays for individual species, especially for those low in demand. A significant amount of resources are saved by having a platform that can be applied to all mammals.
The Horvath Mammalian Array - A Powerful Platform for Comparative Biology and Translational Research
To date, the Horvath Mammalian Array technology has helped to generate accurate epigenetic aging clocks in over 125 mammalian species, and across many different tissues types such as blood, skin, liver, muscle, kidney, brain, oocytes, and many more. The arrays are manufactured by Illumina and include 37k CpG sites that are evolutionarily conserved. It has unique strengths: it applies to all mammalian species even those that have not yet been sequenced, it provides deep coverage of specific cytosines which is a prerequisite for developing robust epigenetic biomarkers, and its focus on highly conserved CpGs increases the chances that findings in one species will translate to those in another species.
The technology allows one to evaluate whether an intervention that affects DNA methylation levels in one species (e.g. mouse, rat, canine, pig) also affects the corresponding DNA methylation levels in another species (e.g. human), and it is tailor-made for cross-species comparisons. In mice for instance, as in humans, there is now an accurate pan-tissue epigenetic clock as well as more than 10 organ or tissue-specific clocks, including for blood, liver, kidney, muscle, heart, fibroblasts, skin, cerebellum, cortex, and more.
Due to its cost-effectiveness, it also enables large scale studies of animal health, e.g. for companion pet health & wellness monitoring and wildlife conservation efforts. Several software tools have been adapted for use with the mammalian methylation array that range from normalization to higher level gene enrichment analysis. Software tools for generating normalized data include SeSaMe and the minfi R package.
© 2025 Clock Foundation.
